My Genome
This page collects genetic information and raw sequence data that I have collected from my own DNA. Feel free to download for non-commercial use, but please acknowledge the source in any publication. Scroll down to find:
- My Y chromosome haplotype
- My mitochondrial sequence
- Genotype data for 500,000 SNPs
- My genome (shallow sequencing)
- My exome
Paternal ancestry by Y-chromosome haplotyping
I know from the written record that my direct paternal ancestors back to the mid-1700s all lived in central Sweden (Västergötland). To find out how they got there, I participated in the Genographic project. Since the Y chromosome is only inherited from your father (and his father, and his father’s father, and his father’s father’s father…), there is an unbroken lineage going back to the first humans in Africa. The result:
Haplogroup I1 (M253)
| DYS393: 13 | DYS439: 11 | DYS388: 14 | DYS385a: 14 |
| DYS19: 15 | DYS389-1: 13 | DYS390: 22 | DYS385b: 15 |
| DYS391: 10 | DYS389-2: 16 | DYS426: 11 | DYS392: 12 |
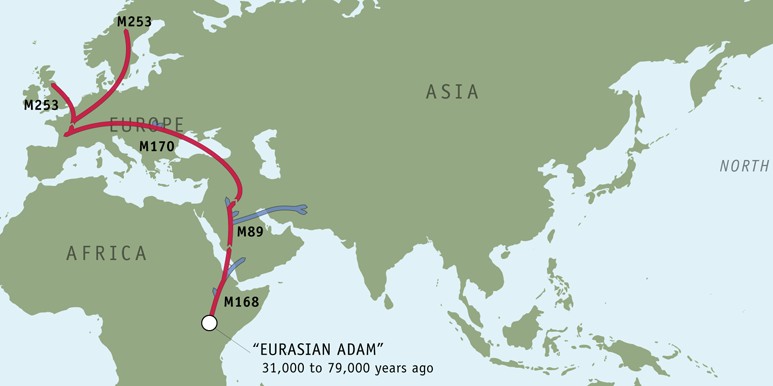
Looks like they took a detour to Bordeax. I wonder what made them leave? Use my personal access code FWJ66EJ3T3 at Genographic to explore the journey!
Maternal ancestry by mitochondrial haplotyping
Since the mitochondrion is only inherited from your mother (and her mother, and her mother…) there is another unbroken lineage going back to Africa through a potentially different route. In fact, every little piece of my genome took a different path from Africa to end up in me!
Anyway, in order to find out where my maternal ancestors came from, I sequenced my mitochondrion. It’s only 16,569 nucleotides (or about five parts per million of the entire genome) so it shouldn’t be too difficult to do nowadays. My sister (who is a nurse) was kind enough to draw some blood from me, and from this I extracted my genomic DNA.
To determine my mitochondrial genome sequence, I aligned about 25,000 reads of 100 bp each against the Cambridge Reference Sequence (CRS) using Mosaik and then basecalled using samtools ‘pileup’. There is nothing special about the CRS other than that geneticists have decided to use it as the reference to which all other mitochondria are compared. Since the genome is so small, I then manually curated each call until I came up with a high-quality list of mutations.
My genotype was:
A73G A263G T282C 309.1C 315.1C 573.1C A750G A1438G A1811G A2706G C3738T A4129G A4769G A5240G T6278G T6392C C6455T C7028T A7055G A8860G C9365T T9698C C10733T G11150A A11467G G11719A C12135A A12308G G12372A G13145A C14766T A15244G A15326G T16209C T16342C
The ones in bold are probably unique to my recent ancestry, but the rest are a perfect match to the U8a1a haplotype. It has been found previously only in Basque, Finnish and English persons, and has been suggested to be one of the ancestral lineages of the Basque going back all the way to the Paleolithic. At some point in time, perhaps around 13,000 years ago, populations around the Iberian peninsula probably migrated north as the ice sheet receded.
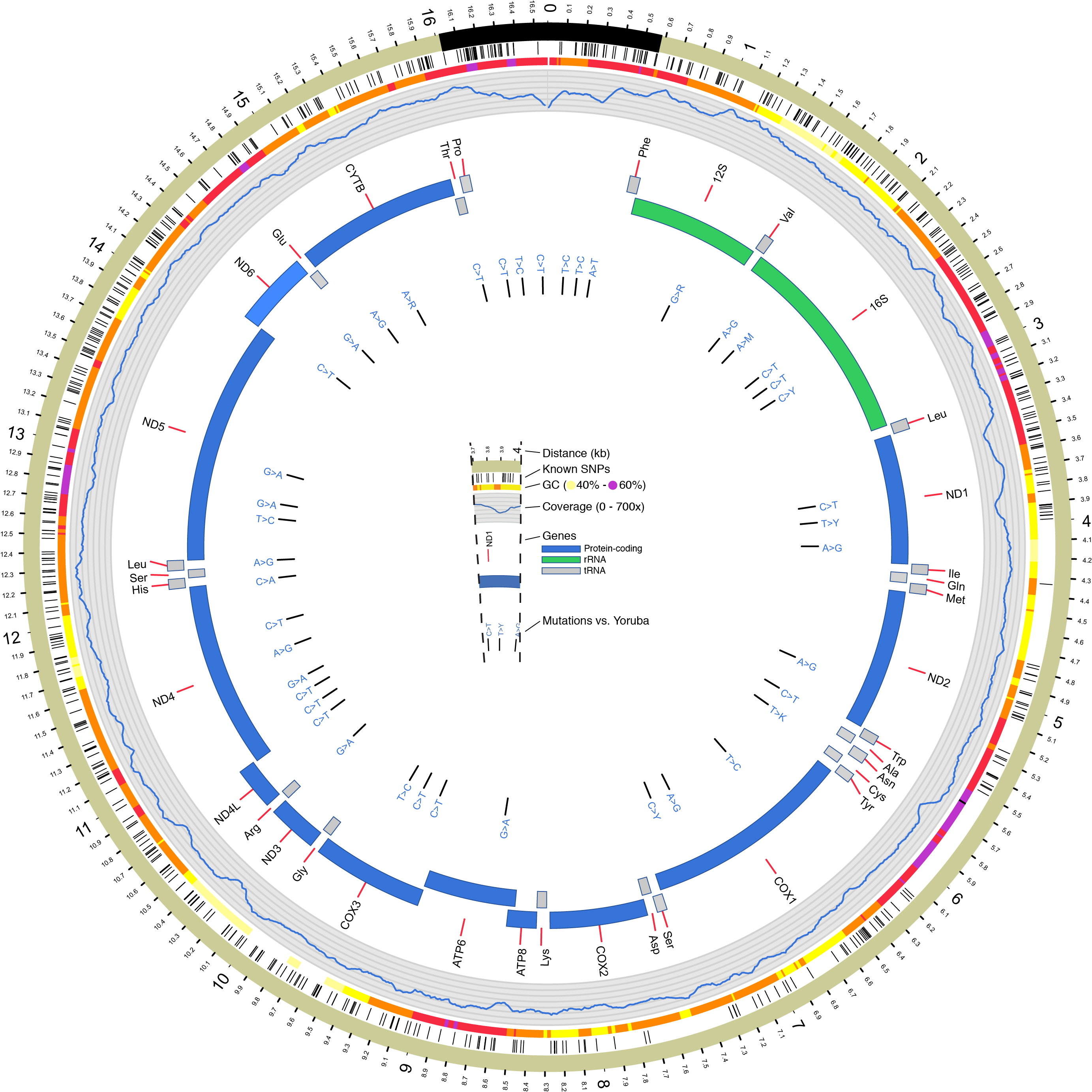
Here’s my mitochondrial genome, with differences from an (ancestral) African mitochondrion indicated:
Affymetrix SNP chip data
To find out more about the origins of my genome, and to see if I carried any disease genes, I genotyped 500,000 single-nucleotide polymorphisms (SNPs). These are small changes in the genome that are usually but not always benign, but can be used as markers to distinguish pieces of the genome and follow their inheritance.
This was then analyzed on the Affymetrix 500K SNP Mapping Array.
Here’s the data: Linnarsson_500k.txt
The dataset was derived from two arrays (StyI and NspI). For convenience, I have combined it into a single file, converted Affy “AB”-type calls into the actual alleles, and replaced Affy SNP ids by standard rs#.
To make sense of the data, I used Promethease to find SNPs associated with disease or other traits. The most important (according to SNPedia) findings were:
- I have an almost two-fold increased risk of coronary heart disease (rs1333049 C/C)
- I have a slightly increased risk of type 2 diabetes (rs7754840 C/C)
A lot of other hits came up, most of which seem dubious or pretty insignificant. I seem to have blue eyes (true!). Two different SNPs suggest that I should be taller than the average person (very true) – but they predict I should be about 0.4 cm taller than average, when it’s more like 20 cm. And I have some SNPs that predict male pattern baldness, although for my age I think I have plenty of hair.