Loompy documentation¶
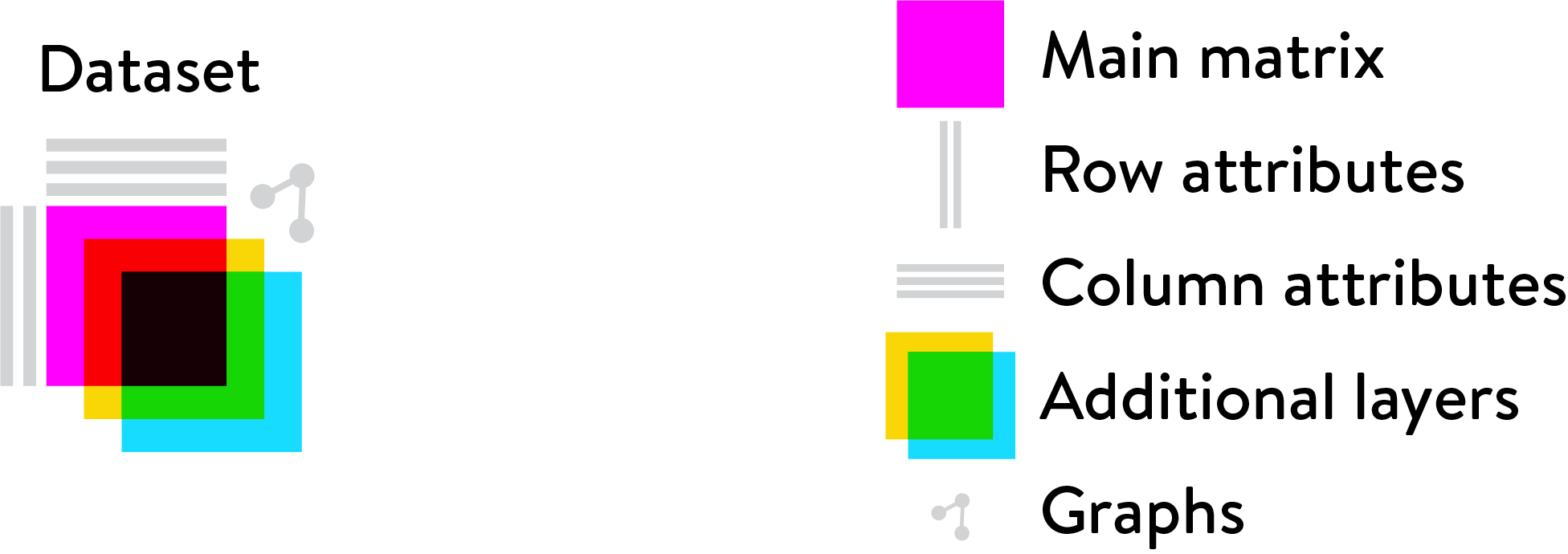
Loom is an efficient file format for very large omics datasets,
consisting of a main matrix, optional additional layers, a variable
number of row and column annotations, and sparse graph objects. We use loom files to store
single-cell gene expression data: the main matrix contains the actual
expression values (one column per cell, one row per gene); row and
column annotations contain metadata for genes and cells, such as
Name, Chromosome, Position (for genes), and Strain,
Sex, Age (for cells). Graph objects are used to store nearest-neighbor
graphs used for graph-based clustering.
The Loom logo illustrates how all the parts fit together:
Loom files (.loom) are created in the
HDF5 file
format, which supports an internal collection of numerical
multidimensional datasets. HDF5 is supported by many computer languages,
including Python,
R,
MATLAB,
Mathematica,
C,
C++,
Java, and
Ruby.